Neue Veröffentlichung in Process Biochemistry: "Experimentally implemented dynamic optogenetic optimization of ATPase expression using knowledge-based and Gaussian-process-supported models”
Sebastián Espinel-Ríos, Gerrich Behrendt, Jasmin Bauer, Bruno Morabito, Johannes Pohlodek, Andrea Schütze, Rolf Findeisen, Katja Bettenbrock, Steffen Klamt
22.05.2024
Experimentally implemented dynamic optogenetic optimization of ATPase expression using knowledge-based and Gaussian-process-supported models
Abstract
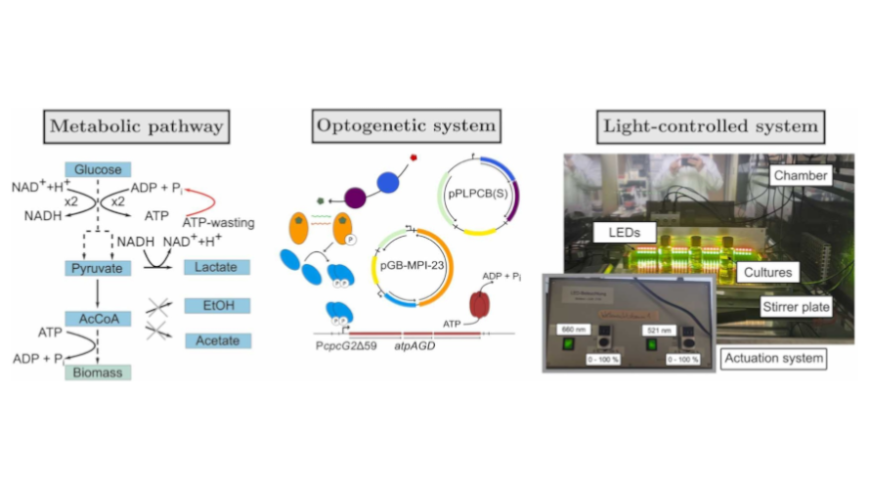
Optogenetic modulation of adenosine triphosphatase (ATPase) expression represents a novel approach to maximize bioprocess efficiency by leveraging enforced adenosine triphosphate (ATP) turnover. In this study, we experimentally implement a model-based open-loop optimization scheme for optogenetic modulation of the expression of ATPase. Increasing the intracellular concentration of ATPase, and thus the level of ATP turnover, in bioprocesses with product synthesis coupled with ATP generation, can lead to increased substrate uptrake and product formation. Previous simulation studies formulated optimal control problems using dynamic constraint-based models to find optimal light inputs in fermentations with optogenetically mediated ATPase expression. However, using these models poses challenges due to resulting bilevel optimizations and complex parameterization. Here, we outline a simplified unsegregated and quasi-unstructured kinetic modeling approach that reduces the number of dynamic states and leads to single-level optimizations. The models can be augmented with Gaussian processes to compensate for model uncertainties. We implement optimal control constrained by knowledge-based and hybrid models for optogenetic ATPase expression in Escherichia coli with lactate as the main product. To do so, we genetically engineer E. coli to obtain optogenetic expression of ATPase using the CcaS/CcaR system. This represents the first experimental implementation of model-based optimization of ATPase expression in bioprocesses.



